Mass Spectrometry After Ip Enrichment
Nearly all Mass spectrometry analysis of PTMs are more effective when combined with IP strategies discussed above to enrich for a protein or PTM of interest. Rather than performing a western blot to determine if a specific protein is modified, the sample is analyzed using a mass spectrometer. The investigator can identify a spectrum of proteins modified by a PTM using bottom-up peptide-based PTM proteomics . Mass spectrometry based PTM identification is a powerful tool, and is a valuable complement to conventional western blot PTM identification especially for site specific identification.
Additional benefits of mass spectrometry are its potentially unbiased approach, and independence from antibodies for detection of PTM modified proteins. While mass spectrometry theoretically provides an unbiased snapshot of PTM modified proteins, it has several limitations which bias against certain types of proteins. Some of the technical challenges include protein abundance bias , and method sensitivity . Fiest and Hummon provide a general review on instrumentation, sample preparation, decontamination, digestion strategies, fractionation approaches, and other considerations when utilizing bottom-up mass spectrometry .
All things considered, initial identification of a specific PTM modified protein using western blot approaches may be easier, require less time, and may be a good first step before utilizing mass spectrometry to address site-specific questions.
Computational Methods For Predicting Ptms
Generally speaking, any computational method for predicting a specific type of PTM has four main steps: data gathering, feature extraction, learning the predictor and performance assessment. These steps have been schematically shown in . In the following, these steps are described in detail. Also, the related challenges and problems in each step are discussed as well.
A schematic flowchart to show how a predictor works for PTM prediction. Data collection and dataset creation. Feature selection. Creating training and testing models. Evaluation of the performance of the models.
The 10 Most Studied Ptms
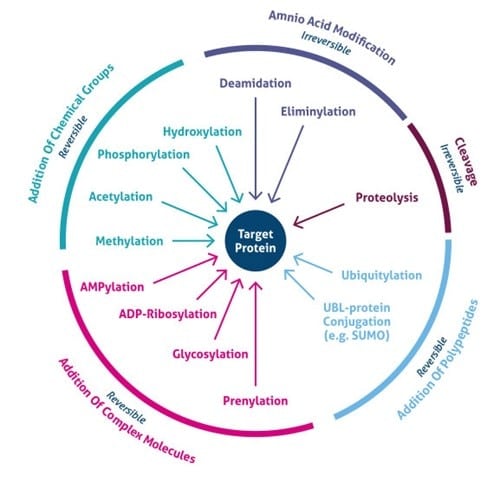
There are more than 400 different types of PTMs affecting many aspects of protein functions. According to the dbPTM , one of the most comprehensive PTM databases, there are 24 major PTMs, with more than 80 experimentally verified reported modified sites. provides a visualized summary of the current major PTM data according to the dbPTM. According to , we can see that some of these major PTMs occur more frequently and have much more been studied. Three main PTMs, based on the dbPTM database, are phosphorylation, acetylation and ubiquitination, which comprise more than 90% of all the reported PTMs Accordingly, each amino acid undergoes at least three different PTMs, and Lys undergoes the largest number of PTMs . Moreover, based on the whole dbPTM data, Cys and Ser are also modified with at least 10 PTM types. Finally, one can see that phosphorylation on Ser is the most reported PTM type.
Summarized information of major PTMs according to the dbPTM databank . All frequencies are shown in log scale. Clustergram indicating the frequency of each PTM on different amino acids. Frequency of major PTMs. Frequency of each amino acid that was reported as a modified site.
Panels B and C in show the frequency of PTM types and amino acids in the dbPTM database in log scale, respectively. According to , it is observed that phosphorylation, acetylation and ubiquitination are the most frequent PTMs.
Recommended Reading: Examples Of High Protein Breakfast
Regulation Of Pluripotency By Protein Acetylation And Hdac Inhibitors
There are five families of HDACs expressed in mammalian cells, including class I , class IIa , class IIb , class III and class IV . Efforts studying HDACs as therapeutic targets in malignant cells have led to the development of a series of small-molecule inhibitors, particularly inhibitors of class I and II HDACs, that block their ability to catalyze protein deacetylation. One of the most well-known HDAC inhibitors is suberoylanilide hydroxamic acid , currently used as an anticancer therapeutic agent to treat patients with cutaneous T-cell lymphoma.
Immunofluorescence For Global Ptm
Immunofluorescence techniques may be a fruitful approach to studying global changes in a PTM profile in tissues or cells. In particular identifying global, and spatial changes in response to drug treament or genetic knockout is achievable with this method. Figure 7 shows example data utilizing Signal-Seeker acetyl lysine antibodies to probe for total acetyl lysine changes. Several Signal-Seeker PTM antibodies have been validated for IF applications.
The identification of target specific PTM modifications are not possible by this method. However, this approach may be applicable as a biological readout thus, may be a useful tool as an indicator for development or disease progression .
Pros: Investigate global PTMs changes in fixed tissue and cells Robust signal May be a useful biological readoutCons: Requires identifying antibodies that are of high IF/IHC quality Requires optimization of fixation techniques No site specificity or protein specificity
Figure 7. Example immunofluorescence data obtained using a pan-acetylation antibody. 3T3 cells were either untreated or treated with TSA for 6 hours. Cells were fixed and stained with pan-acetyl lysine antibody.
Read Also: How To Prevent Gas From Protein Shakes
Overview Of Protein Methylation
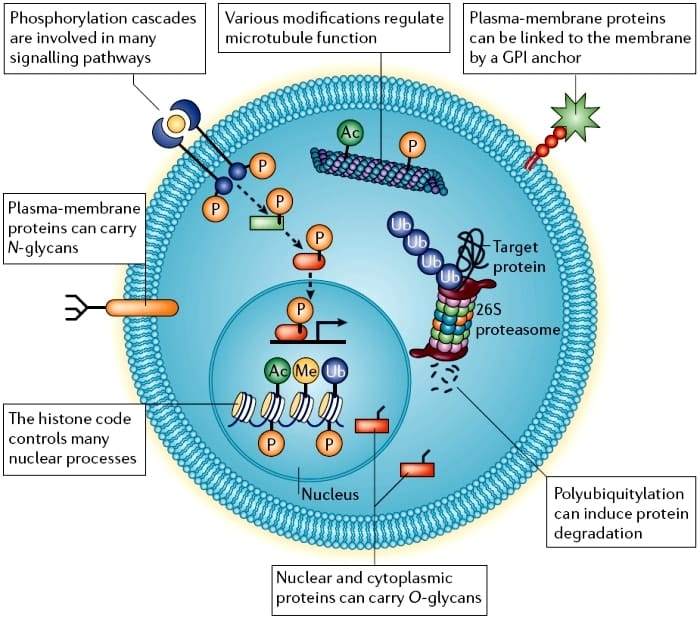
The identity of the enzymes causing protein methylation remained unknown until the heterogeneous nuclear ribonucleoprotein methyltransferase 1 was first discovered in Saccharomyces cerevisiae less than 20 years ago. Since then, numerous types of protein methyltransferases and their orthologs have been identified in yeast, fruit flies and mammals,. It is now clear that protein methylation has profound influences on many biological events and that defects in protein methyltransferases may lead to severe phenotypic abnormalities during embryogenesis,. Two types of protein methylation, arginine and lysine methylation , and their relevant methyltransferases have been frequently described. There are 10 members in the protein arginine methyltransferase family and more than 30 members in the protein lysine methyltransferase family expressed by mammalian cells,. Like HATs, one of the most frequently described substrates for PRMTs and PKMTs is histone. Unlike acetylated lysine residues on histones, which are generally associated with the activation of gene expression, the methylation of different lysine residues on histones may lead to either activation or suppression of gene expression.
Figure 6
In Vitro: Biochemical Assay
Biochemical assays utilize purified or in vitro translated versions of a target protein to determine if it can be modified by a specific PTM. The purified protein is added to a test tube with specific enzymes and the appropriate substrate , co-factors, and energy sources. After incubation, the sample is then analyzed by western blot analysis. Figue 6 provides an example of in vitro ubiquitination of the apoptosis protein BimEL .
It is important to note that in vitro biochemical analysis is not available for all types of PTMs however, in vitro PTM analysis is routinely performed to validate phosphorylation , ubiquitination , SUMOylation and other PTM modifications. A limiting step in performing in vitro biochemical assays is obtaining purified versions of the POI .
Pros: Determine if target protein is modified by specific PTM Useful tool to identify regulatory enzymes for PTM modifitcation of specific POI Good validation tool to confirm PTM modification of target POICons: Proteins, enzymes, and substrates are not at physiologic concentrations, thus, false positive results may be possible. Requires obtaining purified or translated POI as well as enzymes required for specific PTM modification May not be representitive of what occurs in cells and animals
Don’t Miss: Best High Protein Low Carb Bars
Small Changes Huge Impact: The Role Of Protein Posttranslational Modifications In Cellular Homeostasis And Disease
Amrita K. Cheema
Abstract
Posttranslational modifications modulate protein function in most eukaryotes and have a ubiquitous role in diverse range of cellular functions. Identification, characterization, and mapping of these modifications to specific amino acid residues on proteins are critical towards understanding their functional significance in a biological context. The interpretation of proteome data obtained from the high-throughput methods cannot be deciphered unambiguously without a priori knowledge of protein modifications. An in-depth understanding of protein PTMs is important not only for gaining a perception of a wide array of cellular functions but also towards developing drug therapies for many life-threatening diseases like cancer and neurodegenerative disorders. Many of the protein modifications like ubiquitination play a decisive role in various drug response and eventually in disease prognosis. Thus, many commonly observed PTMs are routinely tracked as disease markers while many others are used as molecular targets for developing target-specific therapies. In this paper, we summarize some of the major, well-studied protein alterations and highlight their importance in various chronic diseases and normal development. In addition, other promising minor modifications such as SUMOylation, observed to impact cellular dynamics as well as disease pathology, are mentioned briefly.
1. Introduction
2. Acetylation
3. Carbonylation
4. Glycosylation and Glycation
Regulation Of Pluripotency By Protein Phosphorylation And Dephosphorylation
It is clear that both kinases and phosphatases play critical roles in the proper operation of cell signaling mediated by protein phosphorylation. Unlike many kinases that have been well studied in somatic cells and hPSCs, the importance of protein phosphatases in the regulation of cellular pluripotency is less appreciated. Despite the overwhelming amount of attention that has been focused on kinases in mammalian PSCs, protein phosphatases remain one of the earliest-discovered and most commonly used biomarkers for cellular pluripotency,, indicating the potential functional significance of protein phosphatases in PSCs. Indeed, emerging data have shown that several phosphatases are important for the differentiation capacity and lineage specification of human and murine PSCs. Moreover, suppression of these protein phosphatases inhibits hPSC exit from the pluripotent state during differentiation,,. These studies also illustrate how phosphatases affect cellular pluripotency by altering protein phosphorylation in various signaling pathways, and establish a strong rationale for the development of a strategy to stabilize pluripotency by specific interference with the activity of certain phosphatases.
Recommended Reading: How To Use Protein Shakes To Lose Weight
It All Comes Together In The Er Formation Of Multimeric Proteins
ER is also the place where the subunits come together to create multimeric proteins. Joining of separate polypeptide chains follows the same physical laws as protein folding, an attraction of the hydrophobic surfaces and reactivity of sulfhydryl groups in cysteines. Most often subunits are products of separate genes, and the ER serves as a waiting room for short peptides awaiting longer ones that take a longer time to assemble.
Proteins have to be fully assembled before they are allowed to exit the ER and continue to Golgi.
Pluripotency Is Potentially Affected By Interactions Between Protein Acetylation And Phosphorylation
Like the crosstalk between glycosylation and phosphorylation of proteins, there are many identified interactions between protein acetylation and phosphorylation signaling. For example, the translocation of class IIa HDACs is under the control of Ca2+/calmodulin-dependent protein kinase , cAMP/protein kinase A and protein kinase D -mediated phosphorylation,,,,. The phosphorylation of HDAC3 at Ser424 reduces its deacetylase activity and is antagonistically regulated by casein kinase II and serine/threonine protein phosphatase 4 . The inactivation of SIRT2 through inhibitory phosphorylation at Ser368 is mediated by cyclin-dependent kinases , CDK1, 2 and 5,. HDAC1, 3 and 6 are implicated in the enhancement of AKT signaling through their specific interactions with protein phosphatase 1 and AKT. Moreover, SIRT1 modulates TGF-induced apoptosis by facilitating the deacetylation and degradation of Smad7. These potential regulatory interactions not only add another layer of complexity to the molecular mechanisms underlying cellular pluripotency regulated by protein acetylation, but also remind us that the treatment with HDAC inhibitors that selectively inhibit different types of HDACs may lead to distinct consequences in cellular reprogramming or differentiation of hPSCs.
Don’t Miss: How To Eat Enough Protein
Post Translational Modifications Of Protein
Protein Antibody Specific Ip: Western Blot Analysis
Protein specific IP utilizes an antibody against a POI to immunoprecipitate potentially all species of that protein. The enriched proteins are then separated by SDS-PAGE, transferred to a PVDF membrane, and analyzed via western blot, by probing with a target PTM antibody. Figure 2 provides example data utilizing POI-antibody specific IP, where phosho-SRF was detected using an SRF antibody for IP follwed by phospho serine antibody detection . Target protein antibody IP has traditionally been the first approach used to determine if a POI is modified by a particular PTM, possibly due to the investigator’s expertise with the POI specific antibody for western blot applications.
However, as Figure 1 shows, utilizing the antibody in an IP assay is quite different than its use in a western blot application. It is not surprizing that often times IP with a POI specific antibody fails, or requires significant optimization because it is performing a different function in an IP vs western blot application therefore, as a starting point ensure that the POI specific antibody has been validated for IP. For initial discovery experiments, performing an IP with a PTM specific antibody may be an easier approach, where IP is performed with a PTM specific antibody and the POI specific antibody is used for western blot analysis. This approach only requires optimization of the IP step rather than having to optimize both the IP and the western steps which may be required in a proteinspecific IP.
Also Check: How Much Protein Intake To Gain Muscle
Ptm Antibody Specific Ip: Western Blot Analysis
Figure 2. Example of target protein antibody specific IP. Hippocampal neurons from neonatal pups were grown for 3 d in the presence of either DMSO or 500 nM 6-BIO. Endogenous SRF was immunoprecipitated with anti-SRF antibody and immunoblotted with anti-phosphoserine antibody.
Cong L. Li et al. J. Neurosci. 2014 34:4027-4042
PTM specific IP utilizes an antibody against a target PTM to immunoprecipitate potentially all proteins that have been modified by that PTM . The enriched post-translationally modified population is then separated by SDS-PAGE analysis, transferred to a PVDF membrane, and analyzed via western blot, by probing with an antibody targeting a specific POI.
Figure 3 shows an example of PTM specific IP using tyrosine phosphorylation, ubiquitination, acetylation, and SUMOylation2/3 PTM affinity beads. The EGFR PTM profile of these four modifications was analyzed by probing with an EGFR antibody. This methodology is great for initial discovery of novel PTMs for any POI. PTM specific IP enrichment is also commonly used for bottom-up proteomics , and may be benefical when investigating global PTM changes .
for a detailed protocol for PTM specific IP using Signal-Seeker PTM detection kits.
Pros:
Glycoproteins And Protein Glycosylation
It is well known that protein glycosylation plays a critical role in the regulation of protein structure, signal transduction, cell-cell and cell-environment interactions,,, immune responses,, hormone action, cancer progression and embryonic development,. In the glycosylation process, carbohydrate units can be covalently linked to proteins and edited through various biochemical reactions that are catalyzed by glycosyltransferases and glycosidases in the endoplasmic reticulum and Golgi apparatus . There are four major types of protein glycosylation in mammalian cells: N-linked glycosylation, O-linked glycosylation, C-linked mannosylation and glypiation. Among these types of protein glycosylation, N-linked and O-linked glycomodifications are the most abundant in cells. N-linked glycosylation often occurs on a large variety of nascent proteins. O-linked monosaccharide modification of N-acetylglucosamine on serine, threonine or amino acid residues in close proximity to tyrosine phosphorylation sites is frequently observed in many cells. At these sites, glycosylation may contribute to the regulation of signaling pathways through a direct competition with serine and threonine phosphorylation or by indirectly perturbing the phosphorylation of tyrosine.
Figure 3
Also Check: Eat Your Weight In Protein
Studying A Protein Modification Process In Worms Provides Potential Insights For Human Health
Too much of this post-translational modification impacted multiple body systems
Organisms respond to their environment by changing how several proteins look and interact. These changes, which are called post-translational modifications, are important in the regulation of most biological processes in health and disease.
In a recent study, a team from the laboratory of Mattias Truttmann, Ph.D., assistant professor in the Department of Molecular and Integrative Physiology examined the role of a new post-translational modification called AMPylation, using the round worm Caenorhabditis elegan.
They found that AMPylation is important to control the function of a group of proteins called Transforming Growth Factor- in the worms neurons. They also showed that too much AMPylation stops the growth of worms, impacts reproduction, and blunts their sense of smell.
Like humans, worms rely on their sense of smell to obtain information about their environment. Worms that showed high levels of AMPylation were no longer able to avoid harmful food sources.
Importantly, AMPylation is also happening in humans and TGF- proteins are involved in several human diseases, including cancer and dementia. By discovering how AMPylation controls TGF- proteins in worms, we can thus learn new information relevant to human health.
Paper cited: The AMPylase FIC-1 modulates TGF- signaling in Caenorhabditis elegans, Frontiers in Molecular Neuroscience. DOI: 10.3389/fnmol.2022.912734
Tools For Ptm Prediction
Considering the high cost of experimental identification of PTMs, in recent years, many computational methods have been proposed for the prediction of PTMs. Many of these methods have been introduced as publicly accessible tools. provides a comprehensive list of these tools. In addition to the PTM prediction tools, Nickchi etal. proposed the âPost-translational modification Enrichment Integration and Matching Analysisâ software for carrying out PTM enrichment analysis on proteins . PEIMAN is a publicly accessible standalone software that uses the UniProtKB database to extract PTM terms. In addition to the enrichment analysis, PEIMAN also performs a comparative analysis. In this case, PEIMAN gives two distinct lists of proteins and then integrates the enrichment results and provides a list of highly enriched terms of both protein sets.
Online PTM prediction tools. The values of five important performance assessment measures have been extracted from the related publications: specificity , sensitivity , accuracy , Matthewsâs correlation coefficient and area under the ROC curve .
Don’t Miss: Ka’chava Protein Powder Reviews