Addition Of Chemical Groups
Following translation, small chemical groups can be added onto amino acids within the mature protein structure. Examples of processes which add chemical groups to the target protein include methylation, acetylation and phosphorylation.
Methylation is the reversible addition of a methyl group onto an amino acid catalyzed by methyltransferase enzymes. Methylation occurs on at least 9 of the 20 common amino acids, however, it mainly occurs on the amino acids lysine and arginine. One example of a protein which is commonly methylated is a histone. Histones are proteins found in the nucleus of the cell. DNA is tightly wrapped round histones and held in place by other proteins and interactions between negative charges in the DNA and positive charges on the histone. A highly specific pattern of amino acid methylation on the histone proteins is used to determine which regions of DNA are tightly wound and unable to be transcribed and which regions are loosely wound and able to be transcribed.
Read Also:
Antimicrobial agents are used as protein synthesis inhibitors which include:
The Rna Message Is Decoded On Ribosomes
As we have seen, the synthesis of proteins is guided by information carried by molecules. To maintain the correct and to ensure accuracy , synthesis is performed in the , a catalytic machine made from more than 50 different proteins and several molecules, the ribosomal RNAs . A typical eucaryotic cell contains millions of ribosomes in its . As we have seen, eucaryotic ribosomal subunits are assembled at the , by the association of newly transcribed and modified rRNAs with ribosomal proteins, which have been transported into the after their synthesis in the cytoplasm. The two ribosomal subunits are then exported to the cytoplasm, where they perform protein synthesis.
Ribosomes in the cytoplasm of a eucaryotic cell. This electron micrograph shows a thin section of a small region of cytoplasm. The ribosomes appear as black dots . Some are free in the cytosol others are attached to membranes of the endoplasmic
Eucaryotic and procaryotic ribosomes are very similar in design and function. Both are composed of one large and one small that fit together to form a complete with a mass of several million daltons . The small subunit provides a framework on which the tRNAs can be accurately matched to the codons of the , while the large subunit catalyzes the formation of the peptide bonds that link the amino acids together into a chain .
The Proteasome Degrades A Substantial Fraction Of The Newly Synthesized Proteins In Cells
Cells quickly remove the failures of their translation processes. Recent experiments suggest that as many as one-third of the newly made chains are selected for rapid degradation as a result of the quality control mechanisms just described. The final disposal apparatus in eucaryotes is the , an abundant ATP-dependent protease that constitutes nearly 1% of cellular protein. Present in many copies dispersed throughout the and the , the also targets proteins of the : those proteins that fail either to fold or to be assembled properly after they enter the ER are detected by an ER-based surveillance system that retrotranslocate them back to the cytosol for degradation .
The proteasome. A cut-away view of the structure of the central 20S cylinder, as determined by x-ray crystallography, with the active sites of the proteases indicated by red dots. The structure of the entire proteasome, in which the central cylinder
The 19S caps act as regulated gates at the entrances to the inner proteolytic chamber, being also responsible for binding a targeted to the . With a few exceptions, the proteasomes act on proteins that have been specifically marked for destruction by the covalent attachment of multiple copies of a small protein called . Ubiquitin exists in cells either free or covalently linked to a huge variety of intracellular proteins. For most of these proteins, this tagging by ubiquitin results in their destruction by the proteasome.
Also Check: Whey Protein Powder Weight Loss
Protein Synthesis And Genetic Code
A genetic code or codon is a trinucleotide sequence that identifies a particular amino acid. A codon is a group of three nucleotides so it is called a trinucleotide sequence. For example, a sequence GCC codes for the particular amino acid alanine.
Similarly, 64 codons are present that encode different amino acids. These codons are present in the mRNA template that adjuncts to anticodons present in the tRNA molecule. These anticodons are also trinucleotide sequences.
mRNA, tRNA, and rRNA
There are three types of RNA involved in the process of protein synthesis. The first type is mRNA which carries codons that are converted into an amino acid chain. These mRNA molecules are produced by a DNA template in the nucleus of the cell.
However, in prokaryotes, it synthesizes in the cytoplasm. The mRNA is consists of a 5 cap, 5 UTR region, coding region, 3 UTR region, and a poly chain. The coding region of mRNA is responsible for gene expression which has a start codon at 5 end and a stop codon at 3 end.
Another type of RNA is the tRNA molecule, also called transfer RNA. The transfer of particular amino acid to the ribosome is completed by a tRNA molecule. It has a cloverleaf resembling structure that has two major sites, named as- anticodon arm and accepter stem.
The anticodons are present in the anticodon arm and the accepter stem specifies the particular amino acid that needs to be attached. Now the ribosome translates the mRNA template into a polypeptide chain.
Initiation
Site Of Protein Synthesis
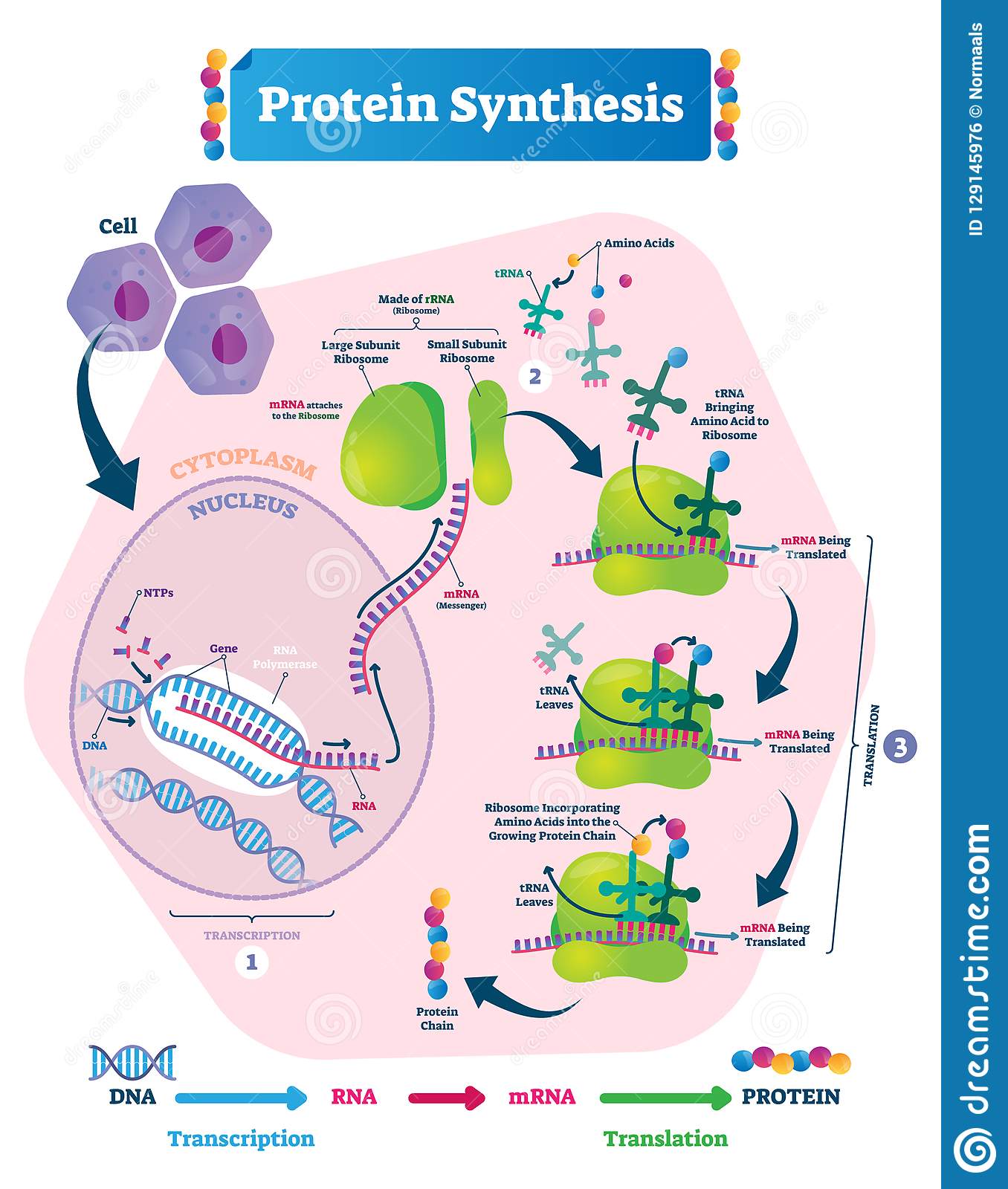
The site of protein synthesis is twofold. Transcription occurs within the cell nucleus where DNA is located. Once the mRNA copy of a small section of DNA has been made it travels through the nuclear pores and into the cell cytoplasm. In the cytoplasm, the strand of mRNA will move towards a free ribosome or one attached to the rough endoplasmic reticulum. Then the next step of protein synthesis translation can begin.
Read Also: Protein Shakes For Picky Eaters
An Mrna Sequence Is Decoded In Sets Of Three Nucleotides
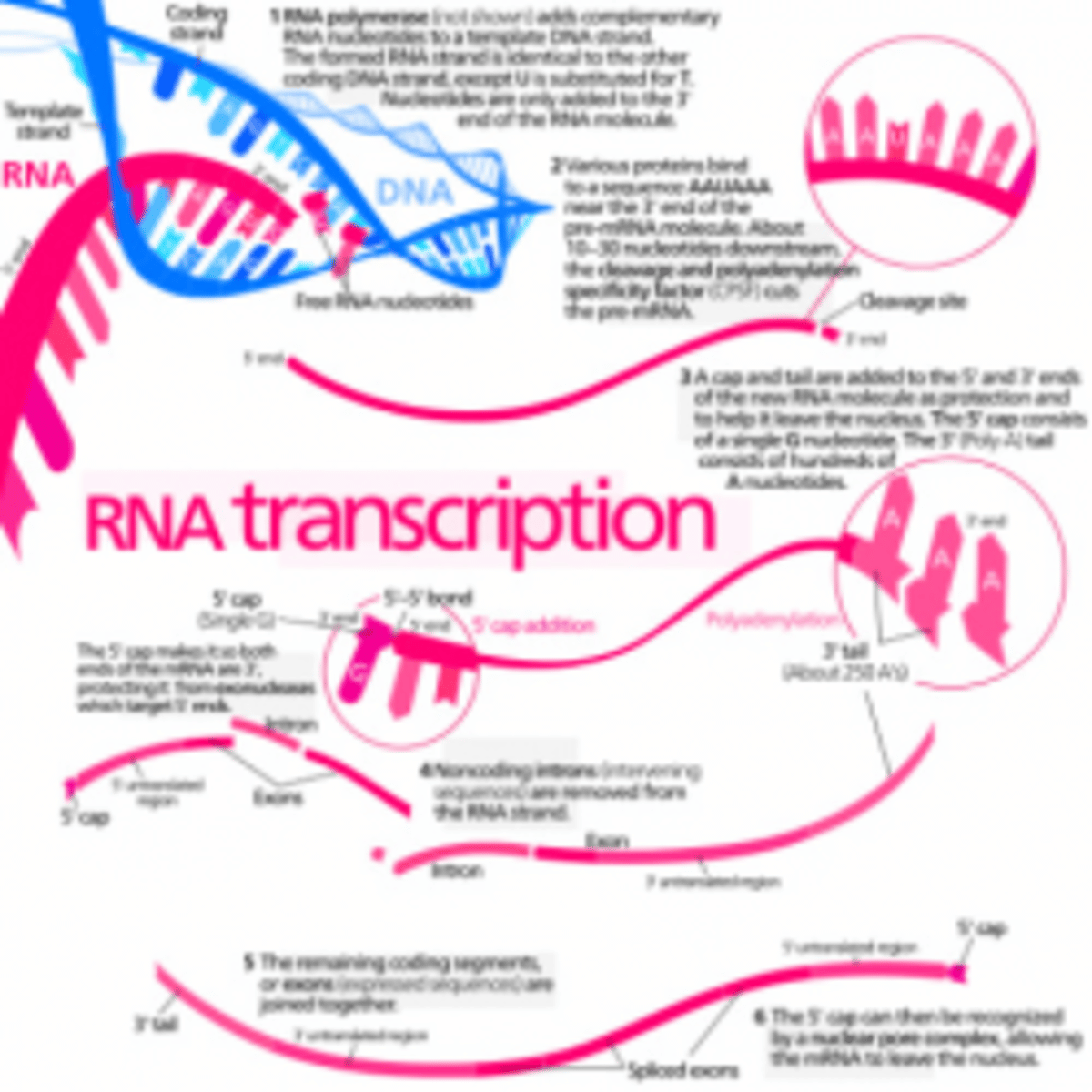
Once an has been produced, by transcription and processing the information present in its sequence is used to synthesize a . Transcription is simple to understand as a means of information transfer: since and are chemically and structurally similar, the DNA can act as a direct for the synthesis of RNA by -pairing. As the term transcription signifies, it is as if a message written out by hand is being converted, say, into a typewritten text. The language itself and the form of the message do not change, and the symbols used are closely related.
In contrast, the conversion of the information in into represents a of the information into another language that uses quite different symbols. Moreover, since there are only four different nucleotides in and twenty different types of amino acids in a protein, this translation cannot be accounted for by a direct one-to-one correspondence between a in RNA and an in protein. The nucleotide sequence of a , through the medium of mRNA, is translated into the amino acid sequence of a protein by rules that are known as the . This code was deciphered in the early 1960s.
The genetic code. The standard one-letter abbreviation for each amino acid is presented below its three-letter abbreviation . By convention, codons are always
Show The People Behind The Science
Storytelling is the most helpful method to grab students attention. Therefore, I spent some time digging a little history of the process to help students relate with the discoverer.
It wont be fair to crown the discovery of protein synthesis to a particular scientist. Zamecnik, a nester of protein synthesis, stated that an impressive list of pioneers blazed the trail to the present scene, making it an eminently multidisciplinary endeavor.
This scientific endeavor dates back to the beginning of the 20th century when Franz Hofmeister and Emil Fischer recognized the peptide bond structure of proteins. Fritz Lipmann enlightened us about the presence of a high-energy phosphate intermediate, while Max Bergmann reported the specificity of proteolytic enzymes. Torbjörn Caspersson and Jean Brachet unraveled the role of RNA in protein synthesis, followed by the discovery of Frederick Sanger, who discovered the first primary protein structure. Sanger also showed the amino acid specification to produce insulin. All this work led up to the efforts of George Palade, who presented visual evidence of protein synthesis with particulate structures in the cytoplasm acting as the cellular sites.
Don’t Miss: How Much Protein To Take
Ubiquitous Influence Of Codons 35 On Protein Synthesis
Fig. 5: The position and context of the motifs around the initiation codon is critical for protein-synthesis yield.
To further examine the effects of codons 35 on protein expression and impact of the 5 non-coding sequences we turned towards vectors that express polycistronic mRNAs. The majority of the E. coli genes are expressed from polycistronic operons where expression of each individual transcript is dependent on ORF-centric structures. We tested expression of the same eGFP variants as in previous experiments, however, now cloned as the second ORF in polycistronic operon with mCherry into two different vectors . In addition, the two vectors had different promoter, intergenic and ribosome binding sequences close to the translation start site. Expression of mCherry reporter was uniform for all constructs regardless of the vector, while eGFP variants were expressed at different levels that correlated with the GFP score determined from our library . Noticeably in vivo expression of eGFP variants from both polycistronic vectors, as well as in vitro expression from T7 polymerase transcribed polycistronic mRNAs, recapitulated data using monocistronic reporter . As such, previously observed differences between eGFP variants were maintained regardless of the polycistronic arrangement, differences in promoter, intergenic sequences or RBS.
Enzymes Involved In Transcription
DNA helicase is the enzyme responsible for the early step of unwinding and unzipping. This enzyme catalyses the breaking of the hydrogen bonds found between complementary base pairs and allows the template strand to be exposed for the next enzyme, RNA polymerase.
RNA polymerase travels along the strand and catalyses the formation of phosphodiester bonds between adjacent RNAnucleotides. Adenine pairs with uracil, while cytosine pairs with guanine.
Remember: in RNA, adenine pairs with uracil. In DNA, adenine pairs with thymine.
You May Like: High Protein Crock Pot Recipes
S Involved In Protein Synthesis Mechanism
ADVERTISEMENTS:
The mechanism of protein synthesis has been studied thoroughly in E.coli. The process of protein synthesis on 70S ribosomes is described in detail below. The summary of the various steps in the mechanism of protein synthesis is shown in .
Step 1 Transcription:
One strand of DNA molecule functions as a template for the formation of mRNA. This mRNA contains the message for the protein to be synthesised. As soon as the mRNA is formed, it leaves the nucleus and reaches the cytoplasm where it attaches with the 30S subunit of ribosomes.
Step 2 Attachment of mRNA to 30S Subunit of Ribosomes:
ADVERTISEMENTS:
In prokaryotic cells, ribosomes are found in a dissociated and an inactive state. The mRNA binds to the 30S subunit. Specific sequences of 16 S rRNA of the small subunit of the ribosome binds to complementary sequence of mRNA. The first amino acid, which is usually N formyl methionine-tRNA attaches to the mRNA to form the initiation complex.
This process is aided by GTP and three protein factors . The ribosome is uniquely designed to bring mRNA and tRNA together. The mRNA is threaded through the small subunit tunnel, while the large and subunit fit together to form tRNA binding pockets. After the formation of the initiation complex, the 50S subunit joins with the smaller subunit to form the 70S ribosome.
Step 3 Transfer of Amino Acids to the Site of Protein Synthesis:
Step 4 Initiation of Protein Synthesis:
Step 5 Elongation of Polypeptide Chain:
Macrolides Chloramphenicol And Oxazolidinones
Protein synthesis, that is, the process of mRNA translation, consists of three steps . These steps involve ribosomes and several other cytoplasmic factors, which play important roles during this process. The ribosome is a complex organelle and is composed of two subunits, the 50 S and 30 S units. Together, they organize the formation of a complex between f-Met-charged-aminoacyl-tRNA and mRNA transcript. This process involves several other initiation factors. Aminoglycosides and tetracycline are one of the most diverse classes of antibiotics , which target this machinery and inhibit protein synthesis in bacteria. Based on the target they choose for inhibitioneither the 50 S inhibitors and the 30 S inhibitorsthey are subdivided into two classes of these antibiotics .
50 S ribosome inhibitors include the macrolide , lincosamide , streptogramin , amphenicol , and oxazolidinone classes of antibiotics. Generally, 50 S ribosome inhibitors bind with it and block either protein translation initiation or peptidyl-tRNAs translocation. This is caused by the inhibition of the peptidyltransferase reaction which is responsible for the elongation of emerging peptide chains .
Alexei A. Sharov, in, 2018
Also Check: How To Make Protein Cookies
Formation Of Covalent Bonds
Many proteins produced within the cell are secreted outside the cell to function as extracellular proteins. Extracellular proteins are exposed to a wide variety of conditions. In order to stabilize the 3D protein structure, covalent bonds are formed either within the protein or between the different polypeptide chains in the quaternary structure. The most prevalent type is a disulfide bond . A disulfide bond is formed between two cysteine amino acids using their side chain chemical groups containing a Sulphur atom, these chemical groups are known as thiol functional groups. Disulfide bonds act to stabilize the pre-existing structure of the protein. Disulfide bonds are formed in an oxidation reaction between two thiol groups and therefore, need an oxidizing environment to react. As a result, disulfide bonds are typically formed in the oxidizing environment of the endoplasmic reticulum catalyzed by enzymes called protein disulfide isomerases. Disulfide bonds are rarely formed in the cytoplasm as it is a reducing environment.
Translation Process In Protein Synthesis
During the translation process, the small and large subunits of a ribosome close over a strand of mRNA, trapping it loosely inside. Ribosomes arrange the strand into codons or sets of three nitrogenous base letters. This is because the code for a single amino acid the most basic form of a protein is a three-letter nucleobase code.
As ribosomes recognize parts of code, we can say they understand it. The jumble of copied letters made during the transcription phase can be read and understood in the translation phase.
For example, GGU, GGC, GGA, and GGG code for the amino acid known as glycine. Most amino acids have multiple codes as this lowers the chance of mistakes if RNA polymerase accidently connects adenine instead of cytosine to GG, it doesnt matter. Both GGC and GGA code for the same amino acid. You can see a list of mRNA codons for the twenty non-essential amino acids here.
There is only one start codon code AUG. Three codons TAA, TAG, and TGA represent stop codons. Neither start nor stop codons match the code for an amino acid they are non-coding. The single start and three stop codons are clearly marked on this codon wheel.
As with the process of transcription, translation within the ribosome is also split into the three stages of initiation, elongation, and termination.
Recommended Reading: What Percent Of Diet Should Be Protein
Specific Enzymes Couple Each Amino Acid To Its Appropriate Trna Molecule
We have seen that, to read the in , cells make a series of different tRNAs. We now consider how each becomes linked to the one in 20 that is its appropriate partner. Recognition and attachment of the correct amino acid depends on enzymes called , which covalently couple each amino acid to its appropriate set of tRNA molecules . For most cells there is a different synthetase for each amino acid one attaches glycine to all tRNAs that recognize codons for glycine, another attaches alanine to all tRNAs that recognize codons for alanine, and so on. Many bacteria, however, have fewer than 20 synthetases, and the same synthetase enzyme is responsible for coupling more than one amino acid to the appropriate tRNAs. In these cases, a single synthetase places the identical amino acid on two different types of tRNAs, only one of which has an that matches the amino acid. A second enzyme then chemically modifies each incorrectly attached amino acid so that it now corresponds to the anticodon displayed by its covalently linked tRNA.
The structure of the aminoacyl-tRNA linkage. The carboxyl end of the amino acid forms an ester bond to ribose. Because the hydrolysis of this ester bond is associated with a large favorable change in free energy, an amino acid held in this way is said
What Are The Four Steps To Protein Synthesis
Steps of Transcription
Also Check: Protein Total Random Urine Low
Activation Of Amino Acids:
Thereaction is brought about when amino acids come to interact with ATP molecules catalyzed by aminoacyl RNA synthetase. The aminoacyl AMP enzyme complex is generated as a result of the reaction between amino acid and adenosine triphosphate , which is mediated by the aforementioned enzyme. The complex is as follows:
AA + ATP Enzyme -AA AMP enzyme complex + PP
Image showing DNA transcription to mRNA
Its worth noting that different amino acids require different aminoacyl RNA synthetases.